Gene Set Enrichment Analysis (GSEA) with clusterProfiler#
require(tidyverse)
Let’s say we have found a set of interesting genes.
geneset_file = file.path(here::here(), "data", "raw", "geneset.txt")
genes_oi = readLines(geneset_file)
genes_oi %>% length() %>% print()
## [1] 166
genes_oi %>% head()
## [1] "ADAM10" "ADAM17" "APAF1" "APBB1" "APH1A" "APOE"
Is is enriched in some biological processes?
require(clusterProfiler)
require(org.Hs.eg.db)
result = enrichGO(genes_oi, ont="BP", keyType="SYMBOL", OrgDb=org.Hs.eg.db)
result %>% as.data.frame() %>% head()
## ID Description
## GO:0006119 GO:0006119 oxidative phosphorylation
## GO:0042775 GO:0042775 mitochondrial ATP synthesis coupled electron transport
## GO:0042773 GO:0042773 ATP synthesis coupled electron transport
## GO:0046034 GO:0046034 ATP metabolic process
## GO:0022904 GO:0022904 respiratory electron transport chain
## GO:0022900 GO:0022900 electron transport chain
## GeneRatio BgRatio pvalue p.adjust qvalue
## GO:0006119 79/157 148/18862 2.595331e-132 7.342192e-129 4.909274e-129
## GO:0042775 62/157 99/18862 1.842964e-108 2.606873e-105 1.743056e-105
## GO:0042773 62/157 100/18862 4.825741e-108 3.501476e-105 2.341222e-105
## GO:0046034 83/157 313/18862 4.950831e-108 3.501476e-105 2.341222e-105
## GO:0022904 63/157 117/18862 3.453413e-104 1.953941e-101 1.306481e-101
## GO:0022900 68/157 178/18862 1.378807e-99 6.501077e-97 4.346872e-97
## geneID
## GO:0006119 ATP5F1A/ATP5F1B/ATP5F1C/ATP5F1D/ATP5F1E/ATP5MC1/ATP5MC2/ATP5MC3/ATP5PB/ATP5PD/ATP5PF/ATP5PO/BID/COX4I1/COX4I2/COX5A/COX5B/COX6A1/COX6A2/COX6B1/COX6B2/COX6C/COX7A1/COX7A2/COX7A2L/COX7B/COX7B2/COX7C/COX8A/COX8C/CYC1/CYCS/NDUFA1/NDUFA10/NDUFA2/NDUFA3/NDUFA4/NDUFA5/NDUFA6/NDUFA7/NDUFA8/NDUFA9/NDUFAB1/NDUFB1/NDUFB10/NDUFB2/NDUFB3/NDUFB4/NDUFB5/NDUFB6/NDUFB7/NDUFB8/NDUFB9/NDUFC1/NDUFC2/NDUFS1/NDUFS2/NDUFS3/NDUFS4/NDUFS5/NDUFS6/NDUFS7/NDUFS8/NDUFV1/NDUFV2/NDUFV3/SDHA/SDHC/SDHD/SNCA/UQCR10/UQCR11/UQCRB/UQCRC1/UQCRC2/UQCRFS1/UQCRH/UQCRHL/UQCRQ
## GO:0042775 BID/COX4I1/COX4I2/COX5A/COX5B/COX6A1/COX6A2/COX6B1/COX6C/COX7A2L/COX7B/COX7C/COX8A/CYC1/CYCS/NDUFA1/NDUFA10/NDUFA2/NDUFA3/NDUFA4/NDUFA5/NDUFA6/NDUFA7/NDUFA8/NDUFA9/NDUFAB1/NDUFB1/NDUFB10/NDUFB2/NDUFB3/NDUFB4/NDUFB5/NDUFB6/NDUFB7/NDUFB8/NDUFB9/NDUFC1/NDUFC2/NDUFS1/NDUFS2/NDUFS3/NDUFS4/NDUFS5/NDUFS6/NDUFS7/NDUFS8/NDUFV1/NDUFV2/NDUFV3/SDHA/SDHC/SDHD/SNCA/UQCR10/UQCR11/UQCRB/UQCRC1/UQCRC2/UQCRFS1/UQCRH/UQCRHL/UQCRQ
## GO:0042773 BID/COX4I1/COX4I2/COX5A/COX5B/COX6A1/COX6A2/COX6B1/COX6C/COX7A2L/COX7B/COX7C/COX8A/CYC1/CYCS/NDUFA1/NDUFA10/NDUFA2/NDUFA3/NDUFA4/NDUFA5/NDUFA6/NDUFA7/NDUFA8/NDUFA9/NDUFAB1/NDUFB1/NDUFB10/NDUFB2/NDUFB3/NDUFB4/NDUFB5/NDUFB6/NDUFB7/NDUFB8/NDUFB9/NDUFC1/NDUFC2/NDUFS1/NDUFS2/NDUFS3/NDUFS4/NDUFS5/NDUFS6/NDUFS7/NDUFS8/NDUFV1/NDUFV2/NDUFV3/SDHA/SDHC/SDHD/SNCA/UQCR10/UQCR11/UQCRB/UQCRC1/UQCRC2/UQCRFS1/UQCRH/UQCRHL/UQCRQ
## GO:0046034 APP/ATP5F1A/ATP5F1B/ATP5F1C/ATP5F1D/ATP5F1E/ATP5MC1/ATP5MC2/ATP5MC3/ATP5PB/ATP5PD/ATP5PF/ATP5PO/BAD/BID/COX4I1/COX4I2/COX5A/COX5B/COX6A1/COX6A2/COX6B1/COX6B2/COX6C/COX7A1/COX7A2/COX7A2L/COX7B/COX7B2/COX7C/COX8A/COX8C/CYC1/CYCS/GAPDH/NDUFA1/NDUFA10/NDUFA2/NDUFA3/NDUFA4/NDUFA5/NDUFA6/NDUFA7/NDUFA8/NDUFA9/NDUFAB1/NDUFB1/NDUFB10/NDUFB2/NDUFB3/NDUFB4/NDUFB5/NDUFB6/NDUFB7/NDUFB8/NDUFB9/NDUFC1/NDUFC2/NDUFS1/NDUFS2/NDUFS3/NDUFS4/NDUFS5/NDUFS6/NDUFS7/NDUFS8/NDUFV1/NDUFV2/NDUFV3/PSEN1/SDHA/SDHC/SDHD/SNCA/UQCR10/UQCR11/UQCRB/UQCRC1/UQCRC2/UQCRFS1/UQCRH/UQCRHL/UQCRQ
## GO:0022904 BID/COX4I1/COX4I2/COX5A/COX5B/COX6A1/COX6A2/COX6B1/COX6C/COX7A2L/COX7B/COX7C/COX8A/CYC1/CYCS/NDUFA1/NDUFA10/NDUFA2/NDUFA3/NDUFA4/NDUFA5/NDUFA6/NDUFA7/NDUFA8/NDUFA9/NDUFAB1/NDUFB1/NDUFB10/NDUFB2/NDUFB3/NDUFB4/NDUFB5/NDUFB6/NDUFB7/NDUFB8/NDUFB9/NDUFC1/NDUFC2/NDUFS1/NDUFS2/NDUFS3/NDUFS4/NDUFS5/NDUFS6/NDUFS7/NDUFS8/NDUFV1/NDUFV2/NDUFV3/SDHA/SDHB/SDHC/SDHD/SNCA/UQCR10/UQCR11/UQCRB/UQCRC1/UQCRC2/UQCRFS1/UQCRH/UQCRHL/UQCRQ
## GO:0022900 BID/COX4I1/COX4I2/COX5A/COX5B/COX6A1/COX6A2/COX6B1/COX6C/COX7A1/COX7A2/COX7A2L/COX7B/COX7B2/COX7C/COX8A/COX8C/CYC1/CYCS/NDUFA1/NDUFA10/NDUFA2/NDUFA3/NDUFA4/NDUFA4L2/NDUFA5/NDUFA6/NDUFA7/NDUFA8/NDUFA9/NDUFAB1/NDUFB1/NDUFB10/NDUFB2/NDUFB3/NDUFB4/NDUFB5/NDUFB6/NDUFB7/NDUFB8/NDUFB9/NDUFC1/NDUFC2/NDUFS1/NDUFS2/NDUFS3/NDUFS4/NDUFS5/NDUFS6/NDUFS7/NDUFS8/NDUFV1/NDUFV2/NDUFV3/SDHA/SDHB/SDHC/SDHD/SNCA/UQCR10/UQCR11/UQCRB/UQCRC1/UQCRC2/UQCRFS1/UQCRH/UQCRHL/UQCRQ
## Count
## GO:0006119 79
## GO:0042775 62
## GO:0042773 62
## GO:0046034 83
## GO:0022904 63
## GO:0022900 68
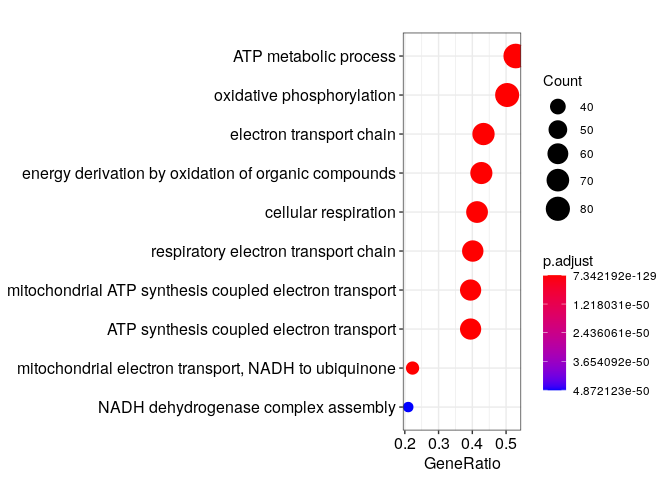
Then, we can quickly get a glimpse of the main results.
result %>% dotplot()
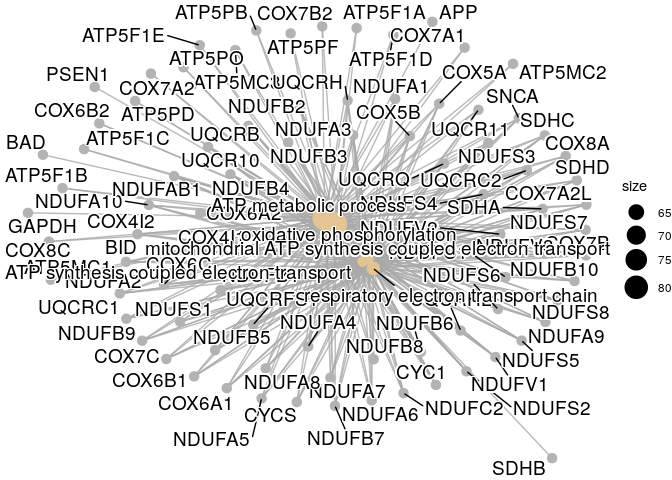
result %>% cnetplot()
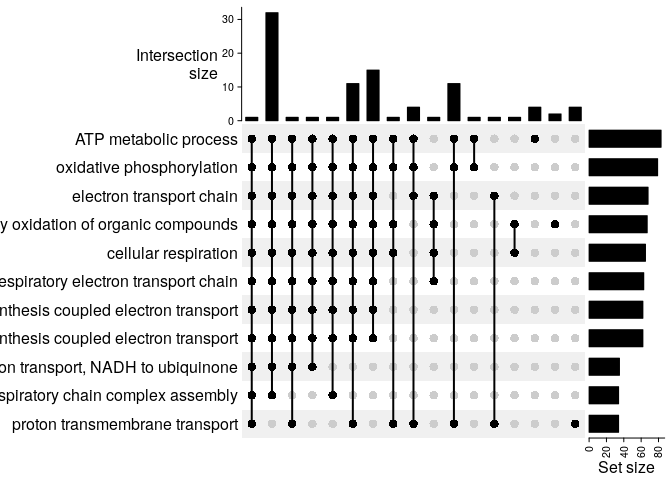
Now, we would like to see the overlap between the genes in the top 5 enriched categories.
require(ComplexHeatmap)
# extract lists of genes in top enriched terms
terms_oi = result %>% as.data.frame() %>% filter(p.adjust<0.05) %>% slice_max(Count, n=10)
terms_oi =
terms_oi %>%
group_by(Description) %>%
summarize(genes=strsplit(geneID, split="/")) %>%
ungroup() %>%
deframe()
# prepare inputs
terms_oi = terms_oi %>%
list_to_matrix() %>%
make_comb_mat()
# plot
plt = terms_oi %>% UpSet()
plt
Phylogenetic trees with ggtree#
Trees are another very common plot to visualize hierarchical patterns.
Here, we will use
ggtree
to visualize the phylogenetic relationships extracted from a multiple
sequence alignment of TP53 across mammals.
Basic tree#
require(ggtree)
set.seed(100)
tree <- rtree(50)
ggtree(tree, layout="circular") + geom_tiplab()
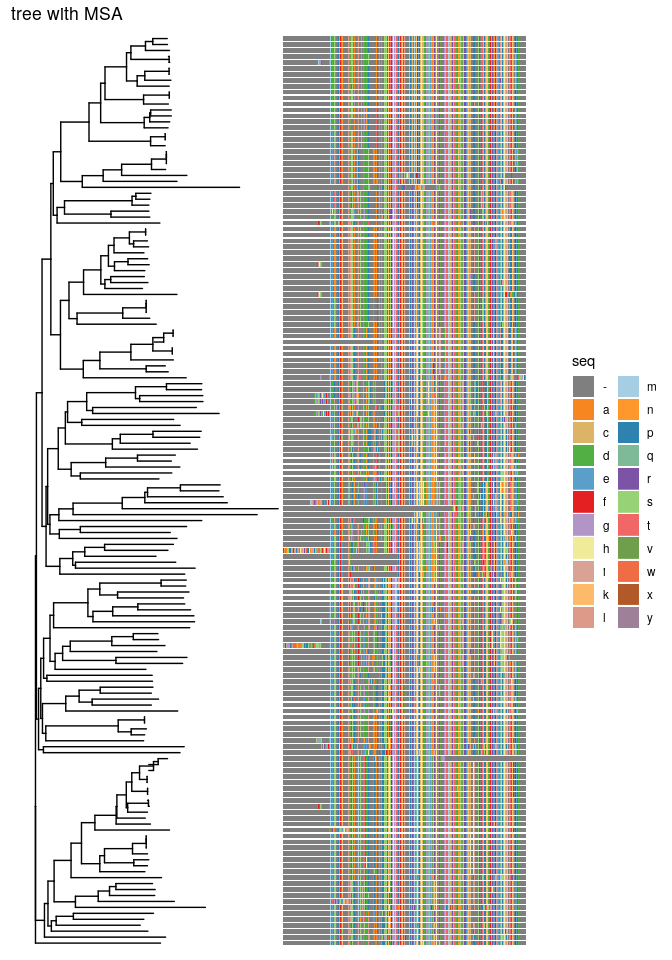
Tree and multiple sequence alignment of TP53 amino acids#
We will follow Russell J. Gray’s approach.
require(seqinr)
require(ape)
# create tree from alignment
fasta_file = file.path(here::here(), "data", "raw", "TP53-mammals-alignment-aa.fa")
aln = read.alignment(fasta_file, format = "fasta", whole.header=TRUE)
D = dist.alignment(aln)
tree = njs(D)
# plot tree with MSA
tree_plot = ggtree(tree)
msaplot(tree_plot, fasta = fasta_file) + ggtitle("tree with MSA")
Session Info#
sessionInfo()
## R version 4.3.1 (2023-06-16)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 18.04.6 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so; LAPACK version 3.7.1
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=es_ES.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=es_ES.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=es_ES.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=es_ES.UTF-8 LC_IDENTIFICATION=C
##
## time zone: Europe/Madrid
## tzcode source: system (glibc)
##
## attached base packages:
## [1] grid stats4 stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] ape_5.7-1 seqinr_4.2-36 ggtree_3.0.4
## [4] ComplexHeatmap_2.9.3 org.Hs.eg.db_3.13.0 AnnotationDbi_1.54.1
## [7] IRanges_2.30.1 S4Vectors_0.40.1 Biobase_2.52.0
## [10] BiocGenerics_0.48.1 clusterProfiler_4.0.5 lubridate_1.9.2
## [13] forcats_1.0.0 stringr_1.5.1 dplyr_1.1.1
## [16] purrr_0.3.4 readr_2.1.5 tidyr_1.2.1
## [19] tibble_3.2.1 ggplot2_3.5.0 tidyverse_2.0.0
##
## loaded via a namespace (and not attached):
## [1] RColorBrewer_1.1-3 shape_1.4.6.1 jsonlite_1.8.0
## [4] magrittr_2.0.3 magick_2.8.3 farver_2.1.1
## [7] rmarkdown_2.26 GlobalOptions_0.1.2 fs_1.6.2
## [10] zlibbioc_1.38.0 vctrs_0.6.1 memoise_2.0.1
## [13] Cairo_1.6-1 RCurl_1.98-1.14 htmltools_0.5.7
## [16] gridGraphics_0.5-1 plyr_1.8.9 cachem_1.0.8
## [19] igraph_1.3.1 lifecycle_1.0.4 iterators_1.0.14
## [22] pkgconfig_2.0.3 Matrix_1.6-5 R6_2.5.1
## [25] fastmap_1.1.1 clue_0.3-65 GenomeInfoDbData_1.2.6
## [28] digest_0.6.29 aplot_0.2.2 enrichplot_1.12.2
## [31] colorspace_2.0-3 patchwork_1.2.0 rprojroot_2.0.4
## [34] RSQLite_2.3.5 labeling_0.4.3 fansi_1.0.3
## [37] timechange_0.3.0 httr_1.4.7 polyclip_1.10-6
## [40] compiler_4.3.1 here_1.0.1 bit64_4.0.5
## [43] withr_3.0.0 doParallel_1.0.17 downloader_0.4
## [46] BiocParallel_1.26.2 viridis_0.6.5 DBI_1.2.2
## [49] highr_0.10 ggforce_0.4.2 MASS_7.3-60
## [52] rjson_0.2.21 tools_4.3.1 DO.db_2.9
## [55] scatterpie_0.2.1 glue_1.6.2 nlme_3.1-164
## [58] GOSemSim_2.18.1 shadowtext_0.1.3 ade4_1.7-22
## [61] cluster_2.1.4 reshape2_1.4.4 fgsea_1.18.0
## [64] generics_0.1.3 gtable_0.3.4 tzdb_0.3.0
## [67] data.table_1.15.2 hms_1.1.3 tidygraph_1.2.0
## [70] utf8_1.2.4 XVector_0.32.0 ggrepel_0.9.5
## [73] foreach_1.5.2 pillar_1.9.0 yulab.utils_0.1.4
## [76] circlize_0.4.16 splines_4.3.1 tweenr_2.0.3
## [79] treeio_1.16.2 lattice_0.21-8 bit_4.0.5
## [82] tidyselect_1.2.1 GO.db_3.13.0 Biostrings_2.64.1
## [85] knitr_1.45 gridExtra_2.3 xfun_0.42
## [88] graphlayouts_1.1.1 matrixStats_1.0.0 stringi_1.7.8
## [91] lazyeval_0.2.2 ggfun_0.1.4 yaml_2.3.8
## [94] evaluate_0.23 codetools_0.2-19 ggraph_2.2.1
## [97] qvalue_2.24.0 ggplotify_0.1.2 cli_3.4.0
## [100] munsell_0.5.0 Rcpp_1.0.8.3 GenomeInfoDb_1.28.4
## [103] png_0.1-8 parallel_4.3.1 blob_1.2.4
## [106] DOSE_3.18.2 bitops_1.0-7 viridisLite_0.4.2
## [109] tidytree_0.4.6 scales_1.3.0 crayon_1.5.2
## [112] GetoptLong_1.0.5 rlang_1.1.0 cowplot_1.1.3
## [115] fastmatch_1.1-4 KEGGREST_1.32.0